The genus Salmonella of the Enterobacteriaceae family comprises two species, Salmonella enterica and Salmonella bongori, both of which are pathogenic for humans and animals. Salmonella enterica spp. is subdivided into 6 subspecies (enterica (I), salamae (II), arizonae (IIIa), diarizonae (IIIb), houtenae (IV) and indica (VI).
The usual habitat for subspecies enterica (I) is warm-blooded animals. The usual habitat for subspecies II, IIIa, IIIb, IV and VI is cold-blooded animals and the environment. All Salmonella can infect humans.
Salmonella strains most commonly isolated from humans and warm-blooded animals belong to S. enterica subsp. enterica. To date, over 2,610 different Salmonella serovars have been described; 59% of these are within S. enterica subsp. enterica I.
Salmonella in humans and livestock
The most commonly reported serovars causing human gastroenteritis infections are S. Enteritidis and S. Typhimurium. The gastrointestinal tract of animals is considered the primary reservoir of the pathogen with human illness usually linked to exposure to contaminated animal-derived products such as meat or poultry.
In 2012, 91,034 human cases of salmonellosis were reported in the EU, a 4.7 % decrease in the number of salmonellosis cases in humans in 2011. A statistically significant decreasing trend in the European Union was observed over the period 2008-2012 mainly a result of the successful Salmonella control programmes in poultry populations.
Typing
Serotyping of Salmonella is based on the antigenic variability of lipopolysaccarides (somatic O antigens) and flagellar proteins (H antigens). The somatic antigens present in the cell wall determine the serogroup and flagellar antigens determine the serovar. The different combinations of the subspecies, 46 O groups, and 114 H antigens, accounts for all recognised serotypes of Salmonella.
Multi-Locus Sequence Type (MLST) analysis based on amplification and sequence interrogation of seven housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA, and thrA) is used as to identify clusters of S. enterica isolates that are related by evolutionary descent. Often sequence types cluster together into discrete, related groups called eBurstGroups (eBGs) and even though not a 1:1 relationship, these have been shown to have good correlation with serogroups1.
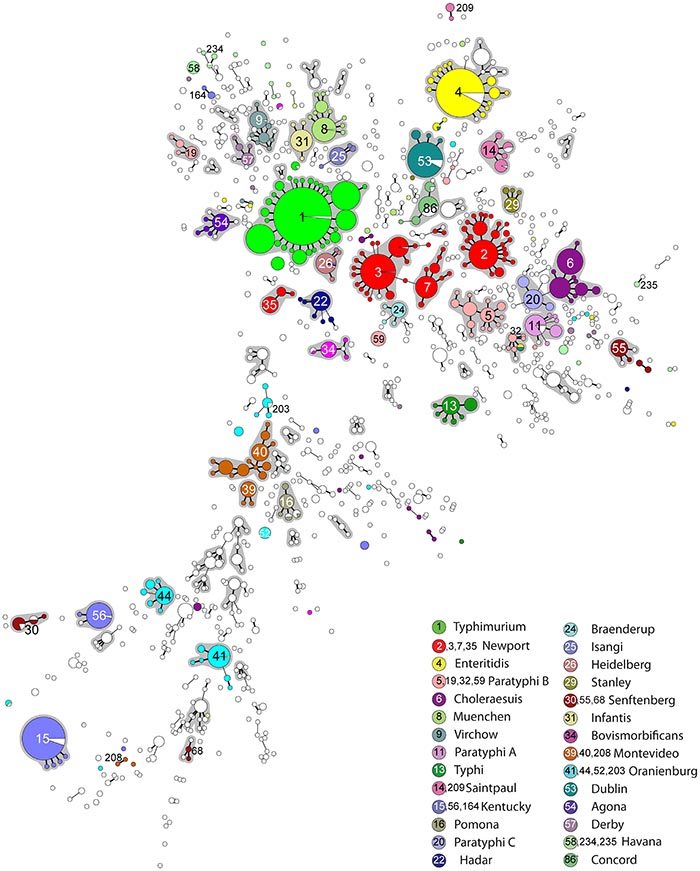
Figure. Minimal spanning tree (MSTree) of MLST data on 4257 isolates of S. enterica subspecies enterica. Each circle corresponds toone of 1,095 STs, whose size is proportional to the number of isolates. The topological arrangement within the MSTree is dictated by its graphic algorithm, which uses an iterative network approach to identify sequential links of increasing distance (fewer shared alleles), beginning with central STs that contain the largest numbers of isolates. As a result, singleton STs are scattered throughout the MSTree proximal to the first node that was encountered with shared alleles, even if equal levels of identity exist to other nodes that are distant within the MSTree. The figure only show links of six identical gene fragments (SLVs; thick black line) and five identical gene fragments (DLVs; thin black line) because these correlate with eBGs, which are indicated by grey shading. The serovar associated with most of the isolates in each eBG or singleton ST is indicated by color coding for the 28 most frequent serovars (see legend at lower right). Within each ST, isolates of a different serovar or for which information is lacking are shown in white, except for monophasic variants.1
doi:10.1371/journal.ppat.1002776.g002
Compare reference set
S. Typhimurium belongs mainly to eBG1 (there are multiple eBGs among S. Typhimurium) and S. Enteritidis to eBG4. The Salmonella reference set consists of 7 publically available genomes to represent 7 of the Salmonella eBGs that contain most frequently isolated Salmonella serotypes from humans and warm-blooded animals:
- S. Typhimurium (eBG1) – reference genome AE006468
- S. Enteritidis (eBG4) – reference genome AM933172
- S. Typhi (eBG13) - reference genome AE014613
- S. Paratypji A (eBG 11) – reference genome CP000026
- S. Paratyphi B/Java (eBG 5) - reference genome CP000886
- S. Agona (eBG54) – reference genome CP001138
- S. Newport (eBG 3) – reference genome CP001113
Last update: July 2015
1Achtman, M. et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 8, e1002776 (2012).