The project for global surveillance of infectious diseases and antimicrobial resistance from sewage has moved into the analytic phase, interpreting the sequencing and residue data of the collected 570 samples from 102 countries.
Please see the updates below.
With support from COMPARE and the World Health Organization, the global sewage surveillance project has moved into the analytic phase interpreting the results.
Update May 2020
In this May 2020 update, we give information about the status of the sampling and analysis that has been undertaken.
The Global Sewage Surveillance Project was initiated in 2016 with the purpose to explore the potential of using sewage for continuous monitoring of AMR. The first results were published in 2019, based on samples collected in 2016 (www.nature.com/articles/s41467-019-08853-3).
All sequence data have been mapped to known databases for antimicrobial resistance genes (ARGs) and bacteria (16S). The abundance of each individual ARG relative to bacterial content in the sample has been calculated. The data have then been analyzed for overall diversity and abundance between the different countries and regions.
Number of countries sampled: 102
Number of samples obtained: 570
For more information, please read the complete Update May 2020.
Sampling is planned to continue for Fall 2020 and Spring 2021.
Update June 2018
In this June 2018 update, we would like to give some information about the status of the pilot run from 2016. The manuscript based on the pilot run from 2016 is still in review. The manuscript is an important step forward acknowledging our proposed surveillance concept.
We have also submitted and made publicly available all of the sequence data from the pilot run - Project acc.:PRJEB27054 / Submission acc.: ERA1502683. We are about to submit the June 2017 sequencing data to the ENA private hub dcc_liszt, which is login protected. This pre-publication, restricted access country-specific data allows the country to perform their own analysis and research on the data. Access can only be granted to the data provider because we want to protect the data until the next publication/ study.
We are of course also continuing with the full-scale urban sewage project, and we know that many of you already anticipate to collect more urban sewage samples in June and November this year. Please note that it is not too late to sign up for June submission if you have missed the opportunity. https://www.surveymonkey.com/r/UrbanSewageSurveillance
Two months ago (April 2018), the Global Sewage Surveillance Project was also adopted by FAO and OIE, which opens up a new phase of the project. This phase (2.2) will run in parallel with the urban sewage collection trials for June and November 2018 (phase 2.1).
In phase 2.2, we want to expand the aim to also include wastewater samples from slaughterhouses for comparison purposes. Slaughterhouses are all very different in design, which is why the wastewater sample should be collected from the slaughter-line including a presentation of as many animals as possible. We want to investigate the interaction and effect between the reservoirs of the healthy animals and healthy humans.
Should you or a colleague of yours, however, also be interested in taking part of this phase 2.2 collection of wastewater samples from slaughterhouses, then please indicate this by signing up.
Please go to the link to express your interest and sign up to receive more information about phase 2.2, slaughterhouse wastewater collection. https://www.surveymonkey.com/r/SlaughterWWS
By comparing sewage from slaughterhouses to the urban sewage samples, we will get a better understanding on how antimicrobial resistance (AMR) genes, pathogens and other factors related to human health may spread from healthy animals over to (healthy) humans through the environment.
Participating institutions/countries will receive access to their sequenced data prior to publication. All individual data-contributors will be acknowledged as a form of co-authorship in the first publication of the full study roll out. The eventual public release of all sequencing data fulfils the aims of open data, open sharing, open science; a common goal among the researchers of this project
We hope all who wish have signed up to participate in this phase 2. If not, there is still time: https://www.surveymonkey.com/r/UrbanSewageSurveillance
Update February 2017
A second phase of sampling for the Global Sewage Surveillance Project has now been opened with the addition of support funding from the Novo Nordisk Fund and continued support from the World Health Organization.
We wish to collect 2L of urban sewage four times in the study period, two times in 2017 and two times in 2018 from major cities around the world. The target is to enroll more than 100 countries and multiple cities within each country.
If you did not participate in the first phase of sampling and wish to join the Global Sewage Surveillance Project, please contact Rene Hendriksen (rshe@food.dtu.dk) for more information about the opportunities from participation.
Update December 2016
The National Food Institute, DTU (WHO Collaborating Centre and European Union Reference Laboratory for Antimicrobial Resistance in Foodborne Pathogens and Genomics) has spent the autumn 2016 interpreting the huge abundance of sequencing data generated. Antimicrobial resistance classes and genes have been identified for each of the samples. Various plots have been created to visually output the abundance profiles for each sample / country (read count matrixes). In conjunction with the identification of the antimicrobial resistance classes and genes, the epidemiological data captured through the survey have similarly undergone analysis categorizing the sample sites into groups.
National Institute for Public Health and the Environment, RIVM (WHO Collaborating Centre for Risk Assessment of Pathogens in Water and Food) has analyzed the samples for antimicrobial residues, which will be the proxy for inadequate global usage data.
Early in 2017, RIVM and DTU will jointly interpret all the data generated, the abundance of AMR genes, the residue data, the epidemiological data, and other health parameters to explain the findings.
Erasmus MC in the Netherlands is still working on sequencing the samples to identify viruses. We anticipate sharing the findings as soon as they become available.
Please contact Rene Hendriksen (rshe@food.dtu.dk) if you are interested to receive the read count abundances of identified resistance genes.
Preliminary results of identified antimicrobial resistance classes and genes for each of the samples.
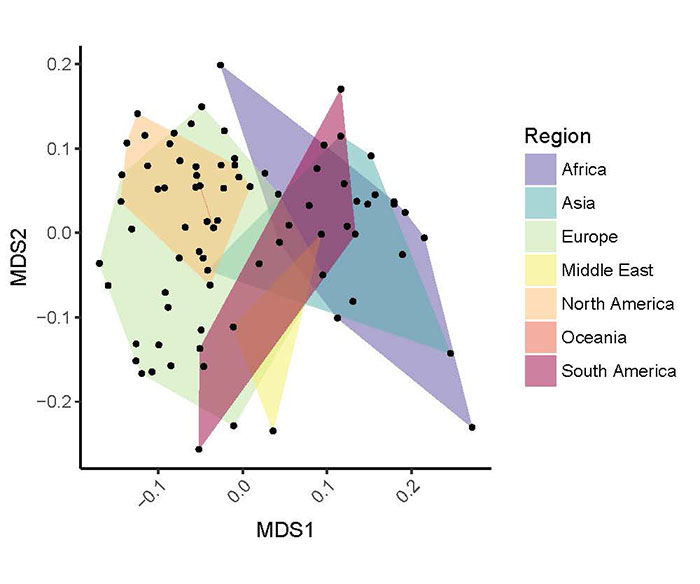
Ordination of Global Sewage resistomes. The read count matrix (genes by samples) was standardized using the Hellinger transformation. The Bray-Curtis dissimilarity indexes between all samples were calculated and principal coordinate analysis (PCoA) was used to project the data so that distances approximate inter-sample dissimilarities. Plotted samples are connected by convex hulls, highlighting geographical regions.
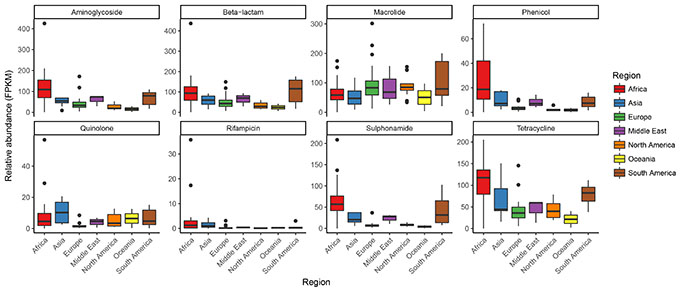
Boxplots of Global Sewage resistance by drug class and region. The read count matrix (genes by samples) was adjusted for gene lengths and sample sequencing depths so that relative abundance (fragments per kilobase reference per million fragments, FPKM) was obtained. Gene abundances within the same drug class were summed and are shown here separated by geographical region.
Preliminary results of identified antimicrobial residues for each of the samples.
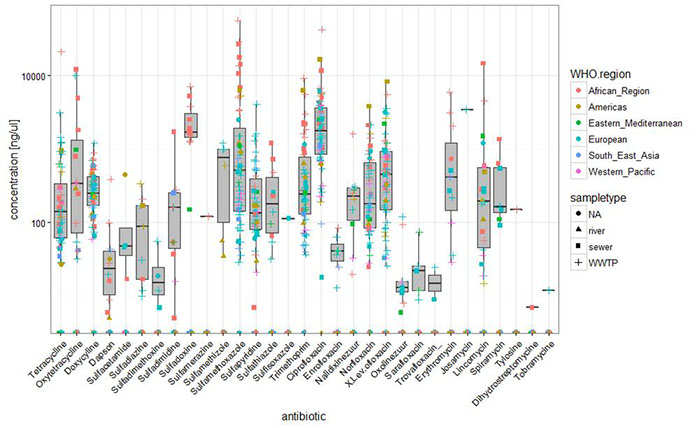
Results: Overall concentrations. This is a log-scale plot of antimicrobial residues. The highest concentrations are found often in open sewers.
Many compounds are not detected, such as beta lactams and cephalosporins, which are degraded before entering the sewers.
General Overview
A video on the project and the process involved is available for viewing.
Metagenomic sequencing of human sewage and quantification of antimicrobial resistance genes and residues combined with epidemiological data is a possible way to determine the occurrence and burden of resistance in defined healthy human populations.
Recent developments in high‐throughput sequencing offer the ability to rapidly identify nucleic acids from various organisms in clinical and environmental samples. Sewage systems are recognized as an important source of human pathogens, especially in crowded settings with poor infrastructure.
The project will serve as proof‐of‐concept for applying metagenomic approaches, which could initiate a global surveillance of human infectious diseases including antimicrobial resistance from sewage collected in major cities around the world to detect, control, prevent and predict human infectious diseases.
Along with The National Food Institute, DTU (WHO Collaborating Centre and European Union Reference Laboratory for Antimicrobial Resistance in Foodborne Pathogens), several other partners from COMPARE are involved in this joint study with WHO, including Erasmus MC, The Netherlands, and National Institute for Public Health and the Environment, RIVM (WHO Collaborating Centre for Risk Assessment of Pathogens in Water and Food).